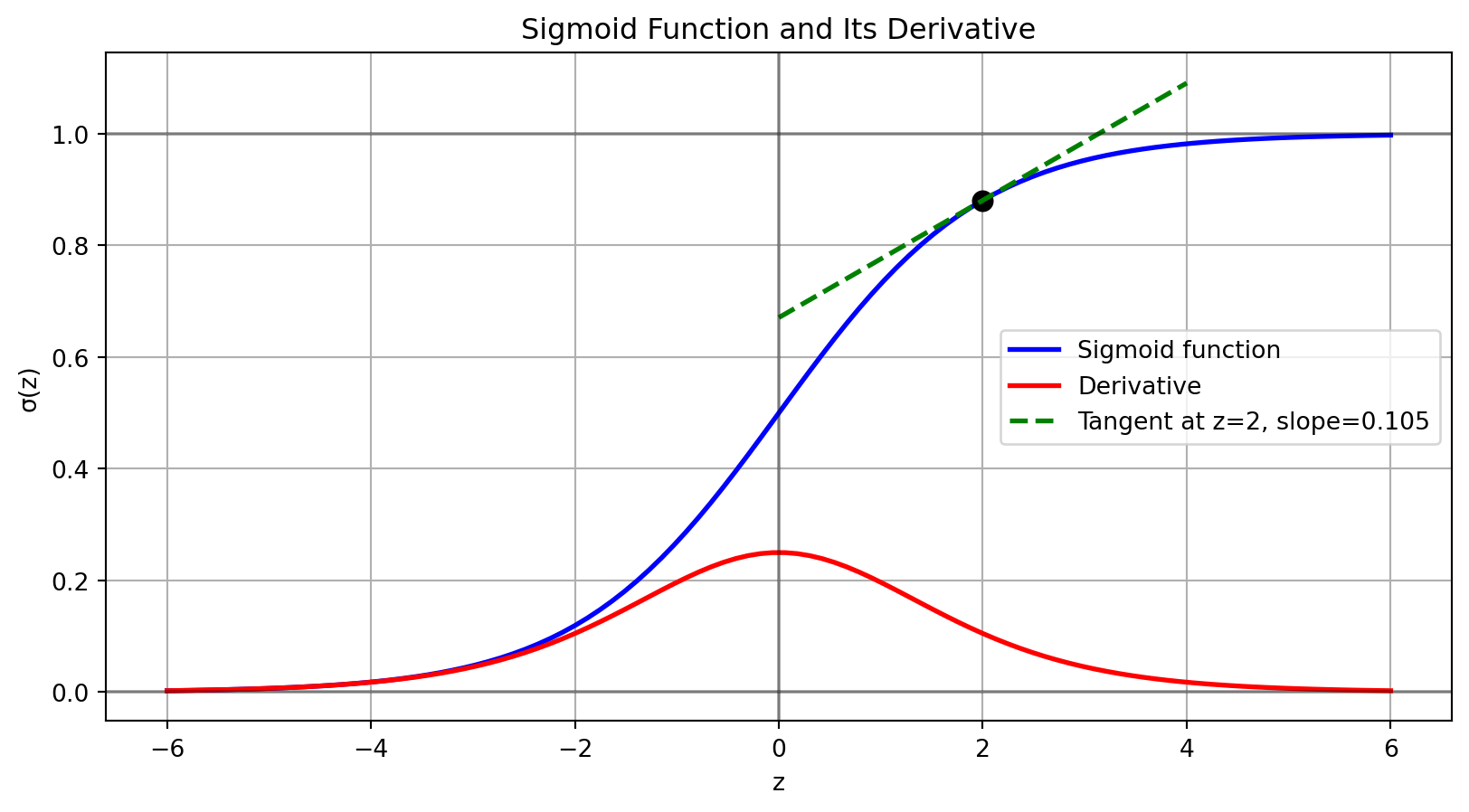
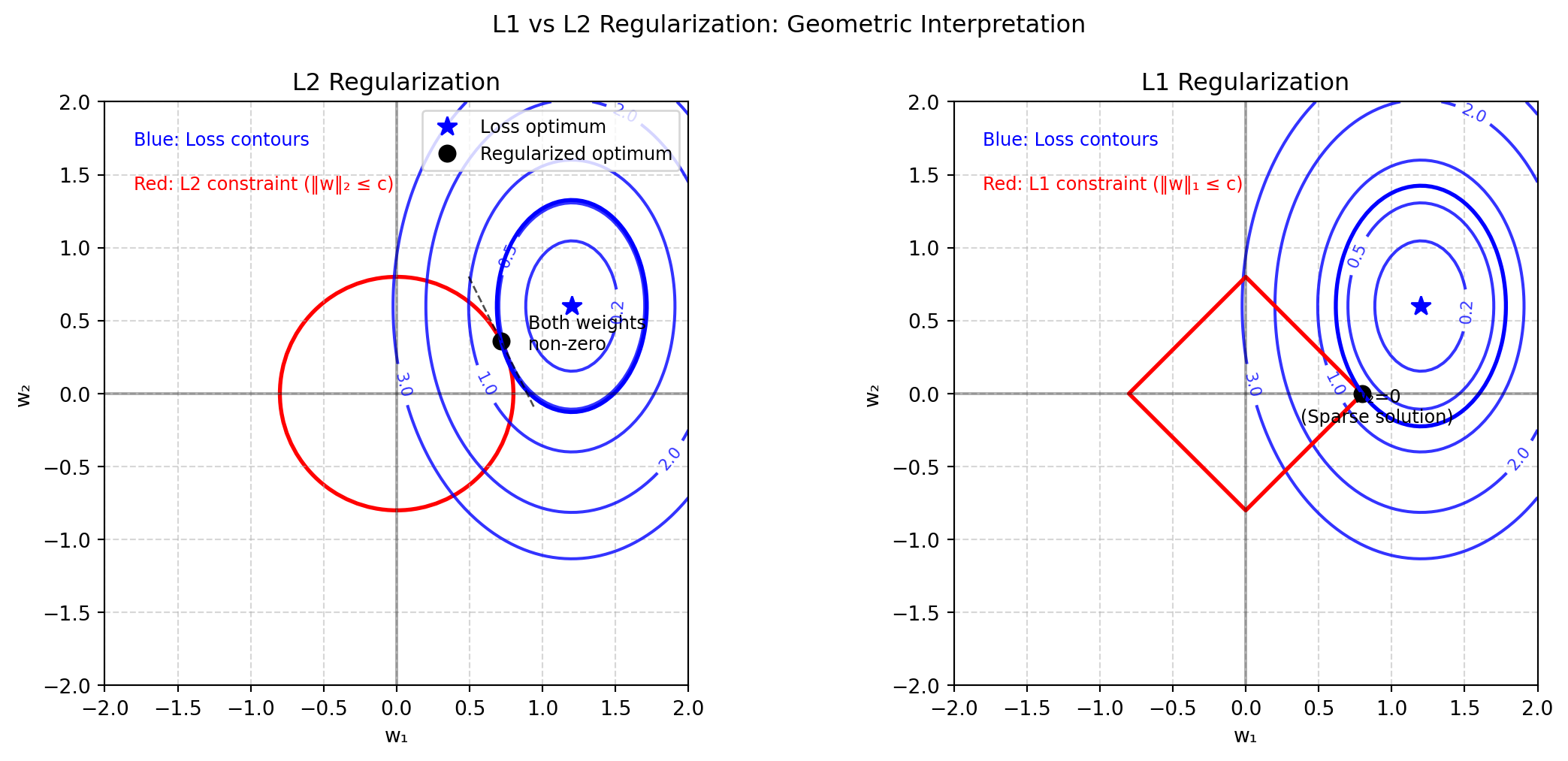
from matplotlib.patches import Circle
# Create geometric visualization of L1 vs L2 regularization
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
# Set up centered coordinates
x = np.linspace(-2, 2, 100)
y = np.linspace(-2, 2, 100)
X, Y = np.meshgrid(x, y)
# True optimal point shifted from origin to show tradeoff
# Modified to be not at 45 degrees from origin for better visualization
optimal_point = np.array([1.2, 0.6]) # Intentionally asymmetric
# Standard loss function (elliptical to avoid 45-degree symmetry)
Z_loss = 2*(X-optimal_point[0])**2 + (Y-optimal_point[1])**2
# Contour levels for loss function
loss_levels = np.array([0.2, 0.5, 1.0, 2.0, 3.0])
# Regularization constraint values
l2_constraint = 0.8 # Radius of circle for L2
l1_constraint = 0.8 # Radius of diamond for L1
# L2 regularization plot
cs0 = ax[0].contour(X, Y, Z_loss, loss_levels, colors='blue', alpha=0.8, linestyles='-')
l2_circle = Circle((0, 0), l2_constraint, fill=False, color='red',
linestyle='-', linewidth=2)
ax[0].add_patch(l2_circle)
# L1 regularization plot
cs1 = ax[1].contour(X, Y, Z_loss, loss_levels, colors='blue', alpha=0.8, linestyles='-')
# L1 diamond shape
l1_points = np.array([
[l1_constraint, 0], [0, l1_constraint],
[-l1_constraint, 0], [0, -l1_constraint], [l1_constraint, 0]
])
ax[1].plot(l1_points[:,0], l1_points[:,1], 'r-', linewidth=2)
# Mark the true optimal point (standard loss only)
for a in ax:
a.plot(optimal_point[0], optimal_point[1], 'b*', markersize=10, label='Loss optimum')
# Analytically determine the constrained optimal points
# For L2: Point on circle closest to optimal point (projection)
l2_direction = optimal_point / np.linalg.norm(optimal_point) # Unit vector toward optimal
l2_optimal = l2_constraint * l2_direction # Scale to lie on circle
# For L1: Point on diamond closest to optimal point
# Due to the asymmetric loss, the optimal point will be on the x-axis
l1_optimal = np.array([l1_constraint, 0]) # Sparse solution with w₂=0
# Plot optimal points
ax[0].plot(l2_optimal[0], l2_optimal[1], 'ko', markersize=8, label='Regularized optimum')
ax[1].plot(l1_optimal[0], l1_optimal[1], 'ko', markersize=8, label='Regularized optimum')
# Calculate loss at optimal points
l2_loss_value = 2*(l2_optimal[0]-optimal_point[0])**2 + (l2_optimal[1]-optimal_point[1])**2
l1_loss_value = 2*(l1_optimal[0]-optimal_point[0])**2 + (l1_optimal[1]-optimal_point[1])**2
# Add custom contours for the exact loss values at optimal points
ax[0].contour(X, Y, Z_loss, [l2_loss_value], colors='blue', linewidths=2)
ax[1].contour(X, Y, Z_loss, [l1_loss_value], colors='blue', linewidths=2)
# Add contour labels
plt.clabel(cs0, inline=1, fontsize=8, fmt='%.1f')
plt.clabel(cs1, inline=1, fontsize=8, fmt='%.1f')
# Labels and formatting
ax[0].set_title('L2 Regularization')
ax[0].text(-1.8, 1.7, 'Blue: Loss contours', color='blue', fontsize=9)
ax[0].text(-1.8, 1.4, 'Red: L2 constraint (‖w‖₂ ≤ c)', color='red', fontsize=9)
ax[0].text(0.9, 0.3, 'Both weights\nnon-zero', color='black', fontsize=9)
ax[0].legend(loc='upper right', fontsize=9)
ax[1].set_title('L1 Regularization')
ax[1].text(-1.8, 1.7, 'Blue: Loss contours', color='blue', fontsize=9)
ax[1].text(-1.8, 1.4, 'Red: L1 constraint (‖w‖₁ ≤ c)', color='red', fontsize=9)
ax[1].text(l1_constraint + 0.1, -0.2, 'w₂=0\n(Sparse solution)', color='black', ha='center', fontsize=9)
# Draw tangent lines at optimal points to emphasize tangency
def get_tangent_points(center, radius, point):
# Get two points on tangent line through optimal point
dx, dy = point[0] - center[0], point[1] - center[1]
norm = np.sqrt(dx**2 + dy**2)
nx, ny = -dy/norm, dx/norm # Normal vector
return [[point[0] - nx*0.5, point[1] - ny*0.5],
[point[0] + nx*0.5, point[1] + ny*0.5]]
# Add tangent line for L2
tangent_l2 = get_tangent_points([0, 0], l2_constraint, l2_optimal)
ax[0].plot([tangent_l2[0][0], tangent_l2[1][0]],
[tangent_l2[0][1], tangent_l2[1][1]],
'k--', linewidth=1, alpha=0.7)
# Set equal axes limits for better comparison
for a in ax:
a.set_xlim(-2, 2)
a.set_ylim(-2, 2)
a.grid(True, linestyle='--', alpha=0.5)
a.set_xlabel('w₁')
a.set_ylabel('w₂')
a.axhline(y=0, color='k', linestyle='-', alpha=0.3)
a.axvline(x=0, color='k', linestyle='-', alpha=0.3)
a.set_aspect('equal') # Equal aspect ratio
plt.tight_layout()
plt.suptitle('L1 vs L2 Regularization: Geometric Interpretation', y=1.05)